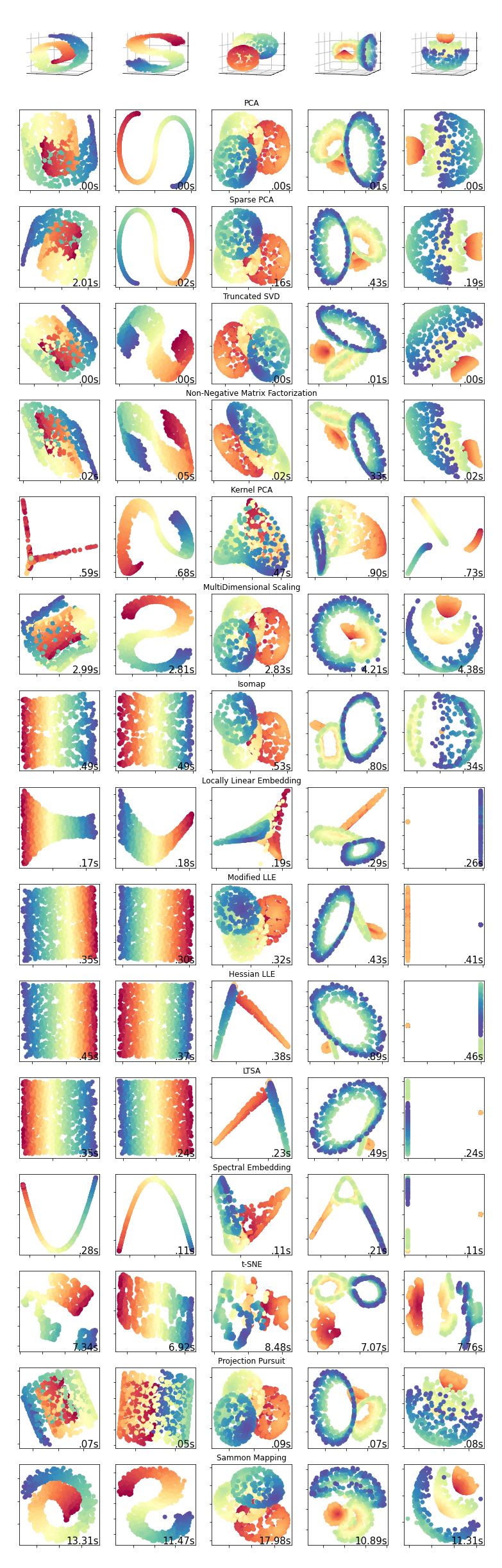
What are the most important Dimensionality Reduction methods in Machine Learning?¶
Dimensionality Reduction is a method for mapping high dimensional inputs into a lower dimension often with the goal preserving most information and hence can be categorized as unsupervised learning.
Here we include a brief summary of important dimensionality reduction methods and a summary chart comparing their results on a set of samples.
Dimensionality Reduction methods can be in general categorized into linear and nonlinear methods. Manifold learning methods may also be considered as nonlinear dimensionality reduction methods.
Linear Dimensionality Reduction
Principal Components Analysis (PCA)
PCA as the most frequently utilized dimensionality reduction, aims to maximize the variance of the data in a lower dimension. This is achieved by identifying the eigenvectors corresponding to the largest eigenvalues.
Sparse Principal Component Analysis (SPCA)
SPCA is a variant of PCA that aims to extract a set of sparse components helping to improve the interpretability of the results.
Truncated Singular Value Decomposition (TSVD) a.k.a. Latent Semantic Analysis (LSA)
TSVD is a variant of SVD that only computes a number of largest singular values and hence reducing the high dimensionality of the data.
Non-Negative Matrix Factorization (NMF)
NMF performs a decomposition of samples matrix by minimizing the distance between the decomposition and the original matrix. Both the samples matrix as well as components contain only non-negative elements.
Nonlinear Dimensionality Reduction
Kernel Principal Component Analysis (KPCA)
Kernel PCA applies the kernel trick to PCA and hence creates a candidate for a nonlinear dimensionality reduction.
Multi Dimensional Scaling (MDS)
Multi Dimensional Scaling aims to arrive at a mapping to a lower dimension with pairwise distances are similar to those in the original data.
Isometric Mapping (Isomap)
Isometric Mapping combines properties from MDS and PCA through three stage algorithm of searching for nearest neighbors, searching for shortest-path graph and finally partial eigenvalue decomposition.
Locally Linear Embedding (LLE)
Locally Linear Embedding aims to obtain a mapping in which the distances are preserved in local neighborhoods and includes three stages of nearest neighbor search, weight matrix construction and partial eigenvalue decomposition.
Modified Locally Linear Embedding (MLLE)
Locally Linear Embedding where multiple weight vectors for various neighborhoods are constructed helping to address the LLE regularization issues.
Hessian Locally Linear Embedding
Hessian LLE applies hessian based quadratic form to recover the locally linear structure and hence helping to address the LLE regularization issues.
Local Tangent Space Alignment (LTSA)
LTSA is similar to LLE method but aims to preserve the local geometry via its tangent space, optimizing the alignments of these local tangent spaces.
Spectral Embedding
Spectral Embedding constructs a weighted graph and its Laplacian and performs a partial eigenvalue decomposition on graph Laplacian. Local distances are expected to be preserved by this method.
t-distributed Stochastic Neighbor Embedding (t-SNE)
t-SNE constructs a probability distribution over the sample assigning probability based on similarity, as well as a probability distribution over a lower dimensional mapping. Finally it optimizes the KL divergence between these distributions with respect to the location of the points in the map.
Projection Pursuit (PP)
Projection Pursuit operates based on initially linearly mapping the input to lower dimension, finding a mapping from this lower dimension to the weighted residuals, and finally constructing the output based on this residual mapping.
Sammon Mapping
Sammon Mapping aims to minimize Sammon’s error which is defined as a function between the bilateral distances between original inputs and their projection.
Comparison
The chart below demonstrates the results of various methods on a number of samples.